Section: New Results
Tree-like Shapes Distance Using the Elastic Shape Analysis Framework
Participants : Alejandro Mottini, Xavier Descombes, Florence Besse.
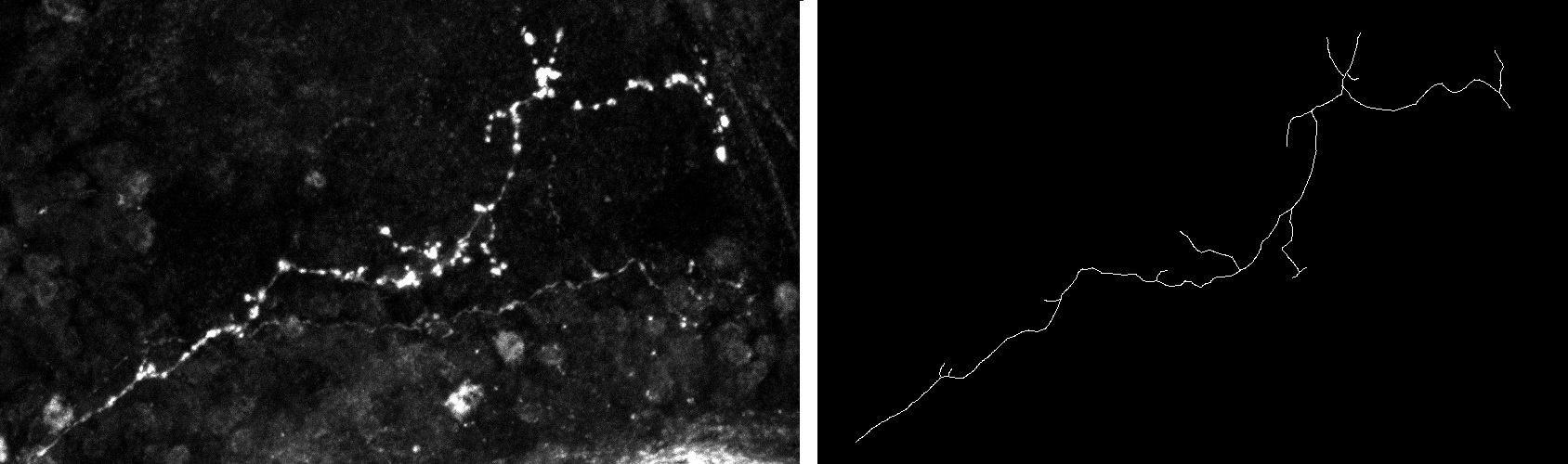
The analysis and comparison of tree-like shapes is of great importance since many structures in nature can be described by them. In the field of biomedical imaging, trees have been used to describe structures such as neurons, blood vessels and lung airways. Since it is known that axon morphology provides information on their functioning and allows the characterization of pathological states, it is of paramount importance to develop methods to analyze their shape and to quantify differences in structures
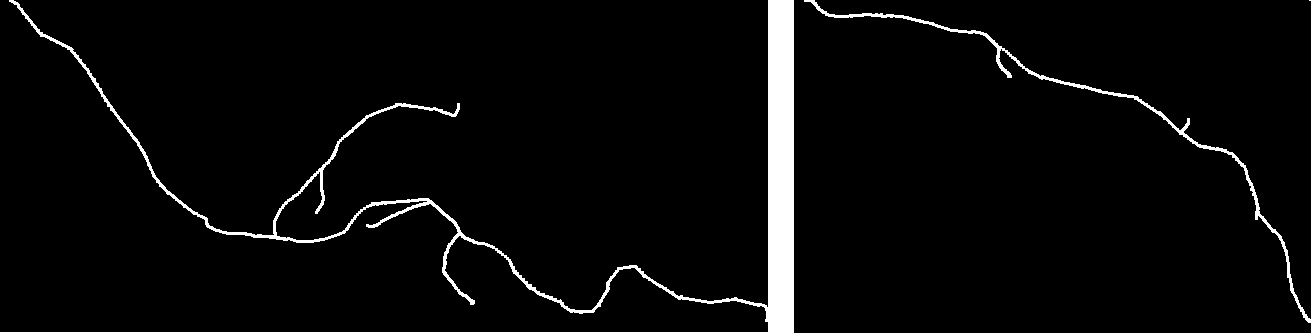
We have developed a new method for comparing tree-like shapes that takes into account both topological and geometrical information [14] , [15] . Our metric combines the Elastic Shape Analysis Framework originally designed for comparing shapes of 3D closed curves in Euclidean spaces with a matching process between branches. Moreover, the method is able to compute the mean shape of a population of trees.
As a first application, we used our method for the comparison of axon morphology. The performance was tested on a group of 61 (20 normal, 24 type one mutant and 17 type two mutant) 3D images, each containing one axonal tree. We have calculated inter and intra class distances between them and implemented a classification scheme. We have compared our results with the ones obtained by three other methods. Results showed that the proposed method better distinguishes between the two populations than the other methods.